Predicting Heart Disease Risk with Random Forest
- William Guesdon
- Mar 1, 2021
- 17 min read
The goal of this project was to predict if a patient present heart disease based on test results, gender and age. The dataset is part of the UCI machine learning dataset.
Photo by Markus Frieauff
Methods
The analysis was performed using R. The dataset was cleaned according to the information gathered on the Kaggle discussion board. I used a decision tree and random Forest algorithms to identify patients with heart disease.
Load libraries
library(tidyverse) # For data cleaning, sorting, and visualisation
library(DataExplorer) # For Exploratory Data Analysis
library(gridExtra) # To plot several plots in one figure
library(ggpubr) # To prepare publication-ready plots
library(GGally) # For correlations
library(caTools) # For classification model
library(rpart) # For classification model
library(rattle) # Plot nicer decision trees
library(randomForest) # For Random Forest modelI Data
heart-disease-uci Kaggle: https://www.kaggle.com/ronitf/heart-disease-uci
Useful links for this dataset
As explained on the links above, it is essential to note that on this dataset, the target value 0 indicates that the patient has heart disease.
Attribute Information:
age: age in years
sex: (1 = male; 0 = female)
cp: chest pain type (typical angina, atypical angina, non-angina, or asymptomatic angina)
trestbps: resting blood pressure (in mm Hg on admission to the hospital)
chol: serum cholestoral in mg/dl
fbs: Fasting blood sugar (< 120 mg/dl or > 120 mg/dl) (1 = true; 0 = false)
restecg: resting electrocardiographic results (normal, ST-T wave abnormality, or left ventricular hypertrophy)
thalach: Max. heart rate achieved during thalium stress test
exang: Exercise induced angina (1 = yes; 0 = no)
oldpeak: ST depression induced by exercise relative to rest
slope: Slope of peak exercise ST segment (0 = upsloping, 1 = flat, or 2 = downsloping)
ca: number of major vessels (0-3) colored by flourosopy 4 = NA
thal: Thalium stress test result 3 = normal; 6 = fixed defect; 7 = reversable defect 0 = NA
target: Heart disease status 1 or 0 (0 = heart disease 1 = asymptomatic)
df <- read_csv("./Data/heart.csv") # To read file on KaggleII Tidy dataset
copy <- df
df2 <- df %>%
filter(
thal != 0 & ca != 4 # remove values correspondind to NA in original dataset
) %>%
# Recode the categorical variables as factors using the dplyr library.
mutate(
sex = case_when(
sex == 0 ~ "female",
sex == 1 ~ "male"
),
fbs = case_when(
fbs == 0 ~ "<=120",
fbs == 1 ~ ">120"
),
exang = case_when(
exang == 0 ~ "no",
exang == 1 ~ "yes"
),
cp = case_when(
cp == 3 ~ "typical angina",
cp == 1 ~ "atypical angina",
cp == 2 ~ "non-anginal",
cp == 0 ~ "asymptomatic angina"
),
restecg = case_when(
restecg == 0 ~ "hypertrophy",
restecg == 1 ~ "normal",
restecg == 2 ~ "wave abnormality"
),
target = case_when(
target == 1 ~ "asymptomatic",
target == 0 ~ "heart-disease"
),
slope = case_when(
slope == 2 ~ "upsloping",
slope == 1 ~ "flat",
slope == 0 ~ "downsloping"
),
thal = case_when(
thal == 1 ~ "fixed defect",
thal == 2 ~ "normal",
thal == 3 ~ "reversable defect"
),
sex = as.factor(sex),
fbs = as.factor(fbs),
exang = as.factor(exang),
cp = as.factor(cp),
slope = as.factor(slope),
ca = as.factor(ca),
thal = as.factor(thal)
)
glimpse(df2) # Check that the transformation worked
plot_missing(df2) # Check that the transformation did not induce NA values
df <- df2 # Replace the df dataset by the tidy datasetIII Exploratory Data Analysis
A Visualise the data summary and distribution of each variable
df %>%
summary()## age sex cp trestbps
## Min. :29.00 female: 95 asymptomatic angina:141 Min. : 94.0
## 1st Qu.:48.00 male :201 atypical angina : 49 1st Qu.:120.0
## Median :56.00 non-anginal : 83 Median :130.0
## Mean :54.52 typical angina : 23 Mean :131.6
## 3rd Qu.:61.00 3rd Qu.:140.0
## Max. :77.00 Max. :200.0
## chol fbs restecg thalach exang
## Min. :126.0 <=120:253 Length:296 Min. : 71.0 no :199
## 1st Qu.:211.0 >120 : 43 Class :character 1st Qu.:133.0 yes: 97
## Median :242.5 Mode :character Median :152.5
## Mean :247.2 Mean :149.6
## 3rd Qu.:275.2 3rd Qu.:166.0
## Max. :564.0 Max. :202.0
## oldpeak slope ca thal
## Min. :0.000 downsloping: 21 0:173 fixed defect : 18
## 1st Qu.:0.000 flat :137 1: 65 normal :163
## Median :0.800 upsloping :138 2: 38 reversable defect:115
## Mean :1.059 3: 20
## 3rd Qu.:1.650
## Max. :6.200
## target
## Length:296
## Class :character
## Mode :character Use the DataExplorer library to get a sense of the distribution of the continuous and categorical variables.
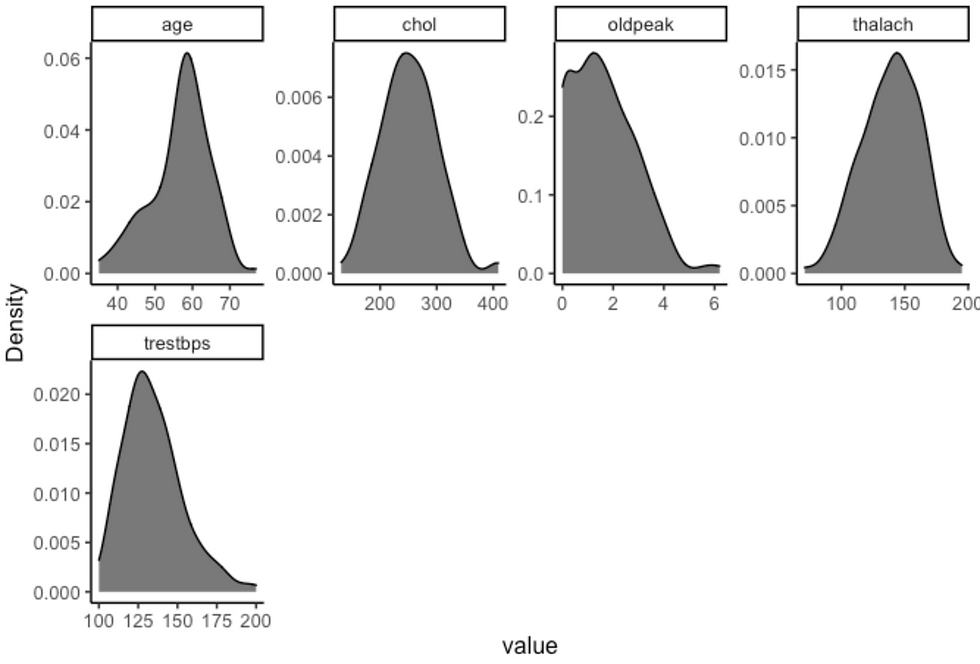
plot_density(df, ggtheme = theme_classic2(), geom_density_args = list("fill" = "black", "alpha" = 0.6))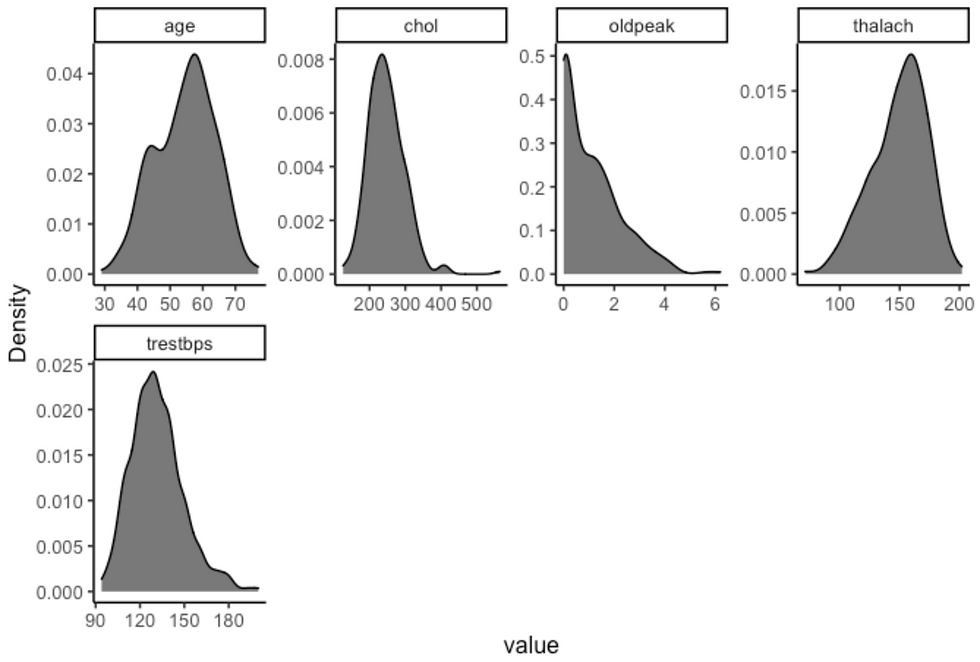
plot_bar(df, ggtheme = theme_classic2())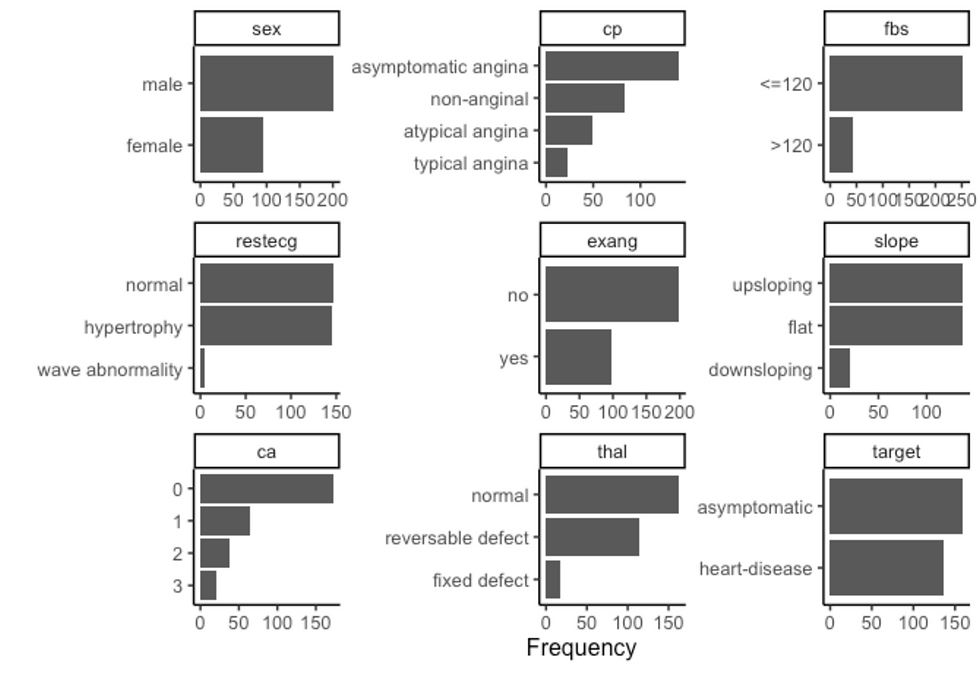
The next step is to combine dplyr and Data Explorer libraries to visualize the variables according to gender and disease.
B Analyse each variable per gender
df %>%
filter(sex == "female") %>%
plot_density(ggtheme = theme_classic2(), geom_density_args = list("fill" = "black", "alpha" = 0.6))
df %>%
filter(sex == "male") %>%
plot_density(ggtheme = theme_classic2(), geom_density_args = list("fill" = "black", "alpha" = 0.6))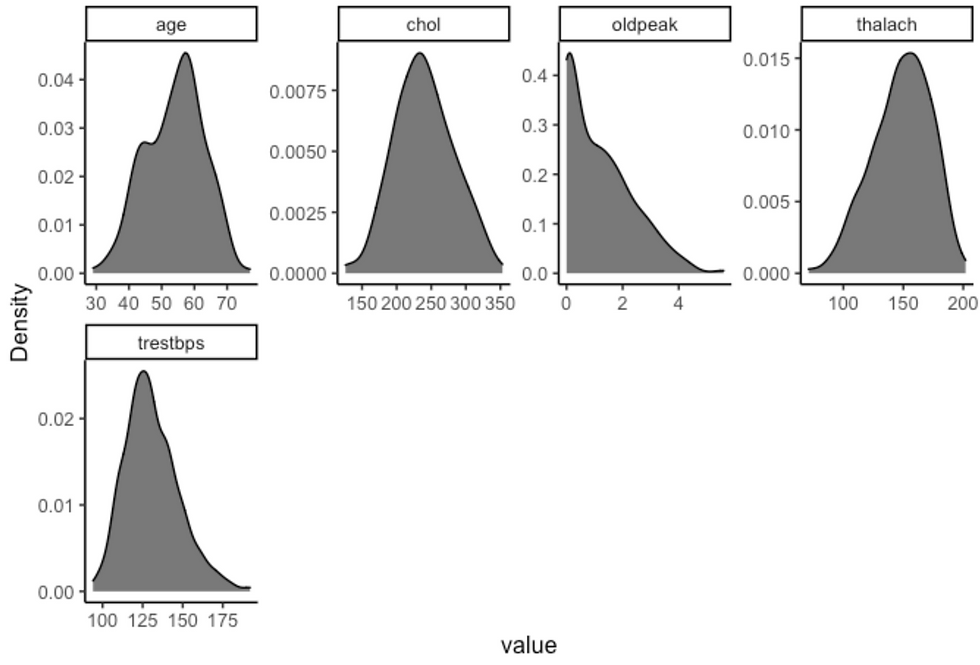
df %>%
filter(sex == "female") %>%
plot_bar(ggtheme = theme_classic2())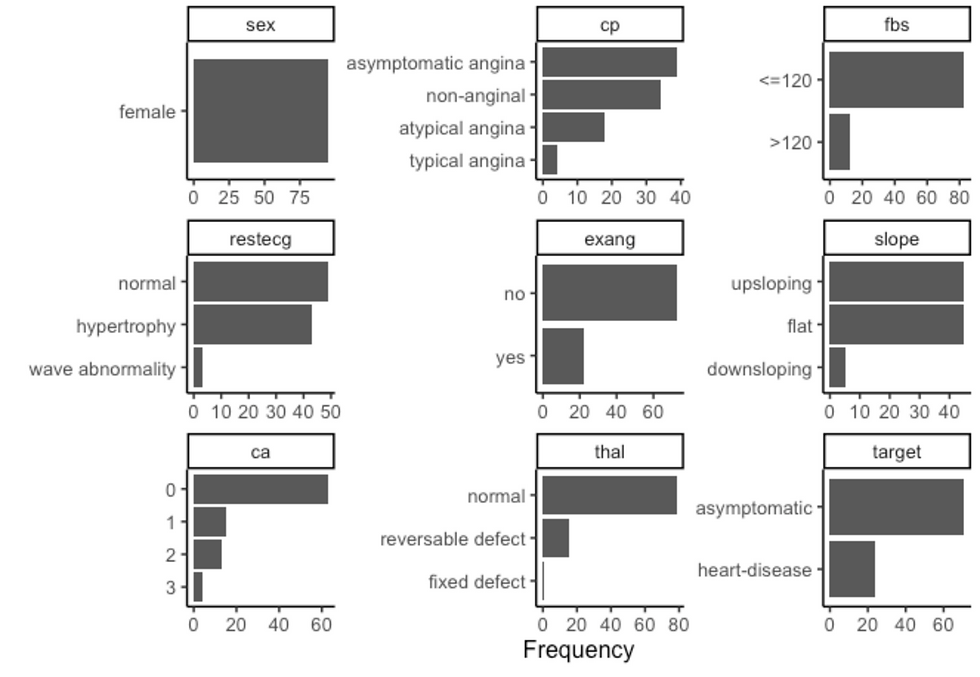
df %>%
filter(sex == "male") %>%
plot_bar(ggtheme = theme_classic2())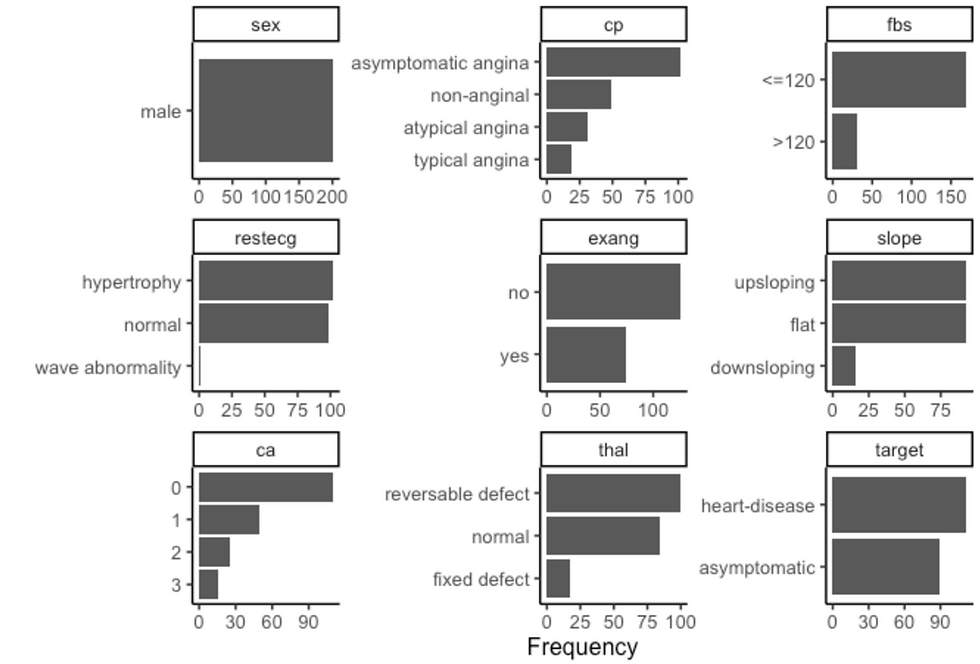
C Visualise variables per disease status
df %>%
filter(target == "asymptomatic") %>%
plot_density(ggtheme = theme_classic2(), geom_density_args = list("fill" = "black", "alpha" = 0.6))
df %>%
filter(target == "heart-disease") %>%
plot_density(ggtheme = theme_classic2(), geom_density_args = list("fill" = "black", "alpha" = 0.6))
df %>%
filter(target == "asymptomatic") %>%
plot_bar(ggtheme = theme_classic2())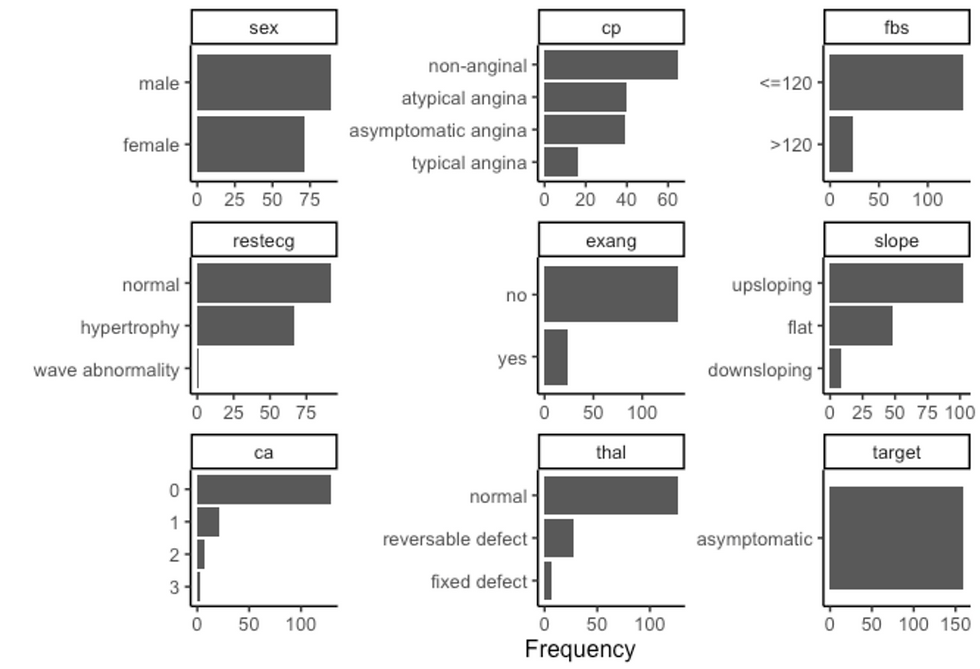
df %>%
filter(target == "heart-disease") %>%
plot_bar(ggtheme = theme_classic2())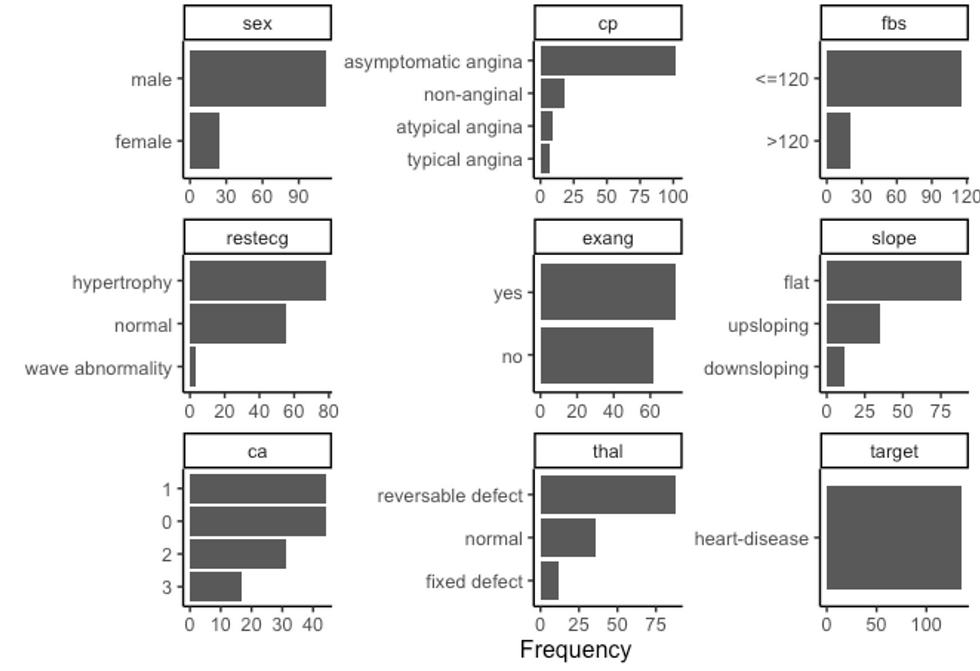
D Visualise the data per gender and disease status
df %>%
filter(sex == "female", target == "asymptomatic") %>%
plot_density(ggtheme = theme_classic2(), geom_density_args = list("fill" = "black", "alpha" = 0.6))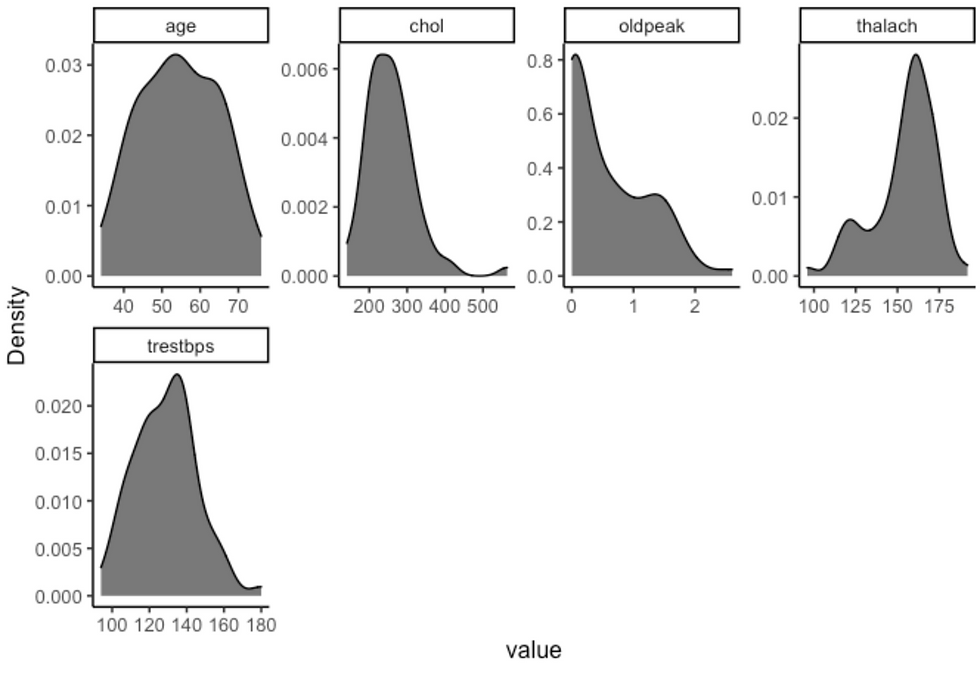
df %>%
filter(sex == "female", target == "heart-disease") %>%
plot_density(ggtheme = theme_classic2(), geom_density_args = list("fill" = "black", "alpha" = 0.6))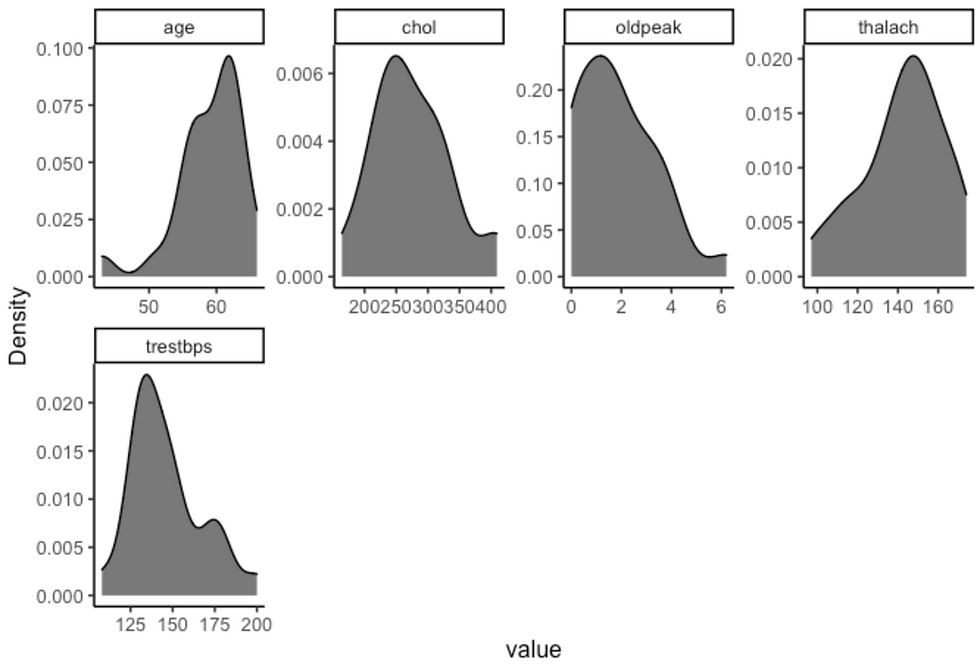
df %>%
filter(sex == "female", target == "asymptomatic") %>%
plot_bar(ggtheme = theme_classic2())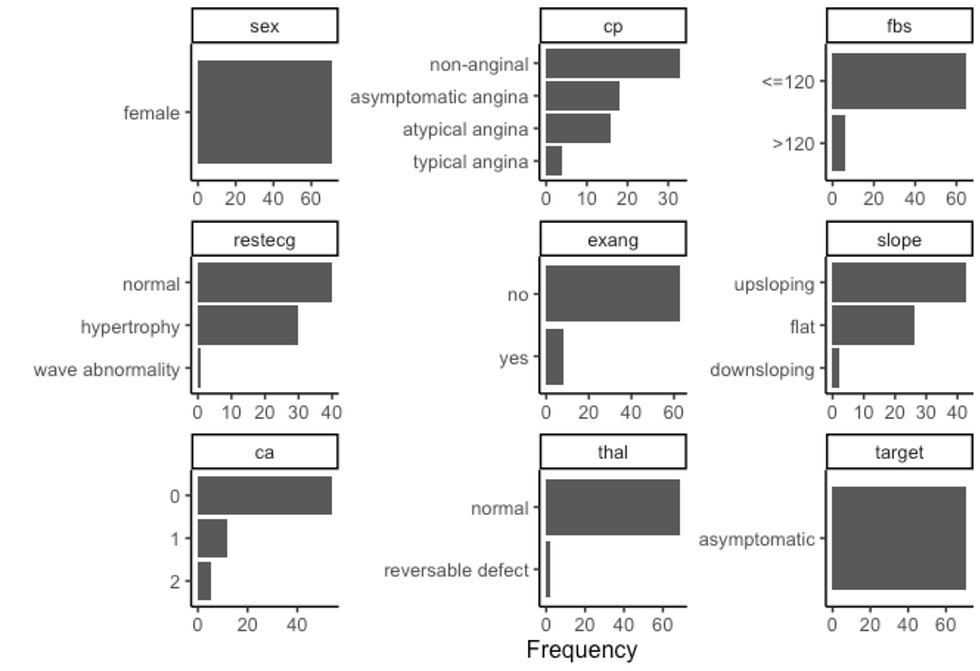
df %>%
filter(sex == "female", target == "heart-disease") %>%
plot_bar(ggtheme = theme_classic2())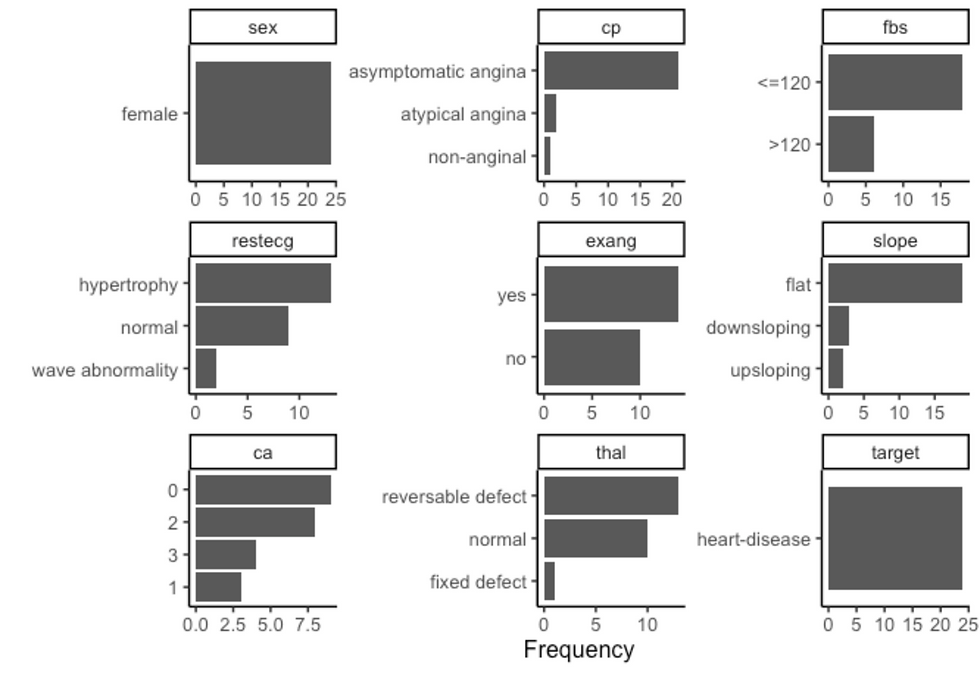
df %>%
filter(sex == "male", target == "asymptomatic") %>%
plot_density(ggtheme = theme_classic2(), geom_density_args = list("fill" = "black", "alpha" = 0.6))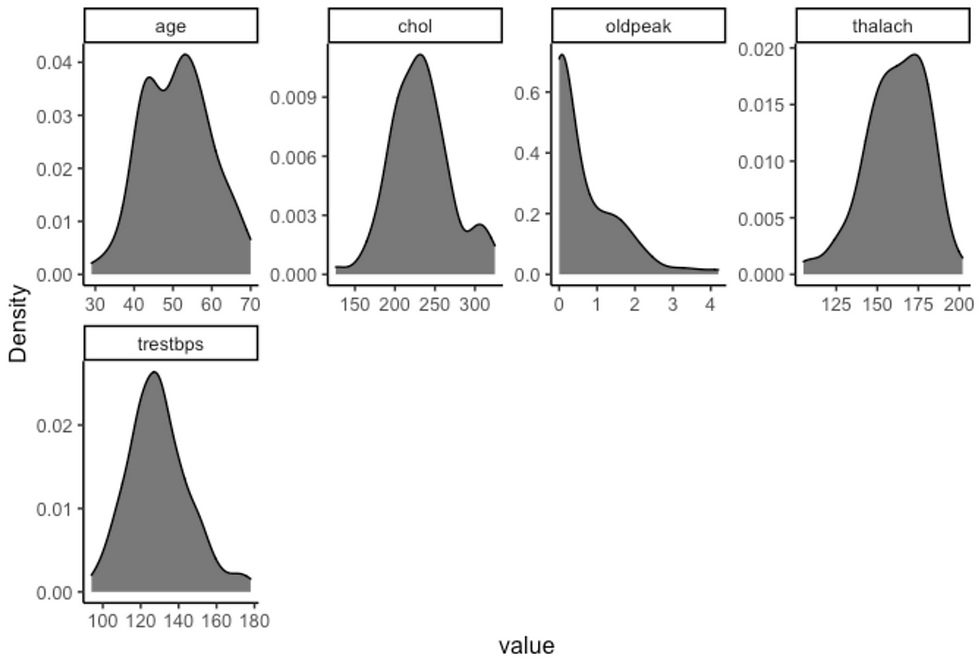
df %>%
filter(sex == "male", target == "heart-disease") %>%
plot_density(ggtheme = theme_classic2(), geom_density_args = list("fill" = "black", "alpha" = 0.6))
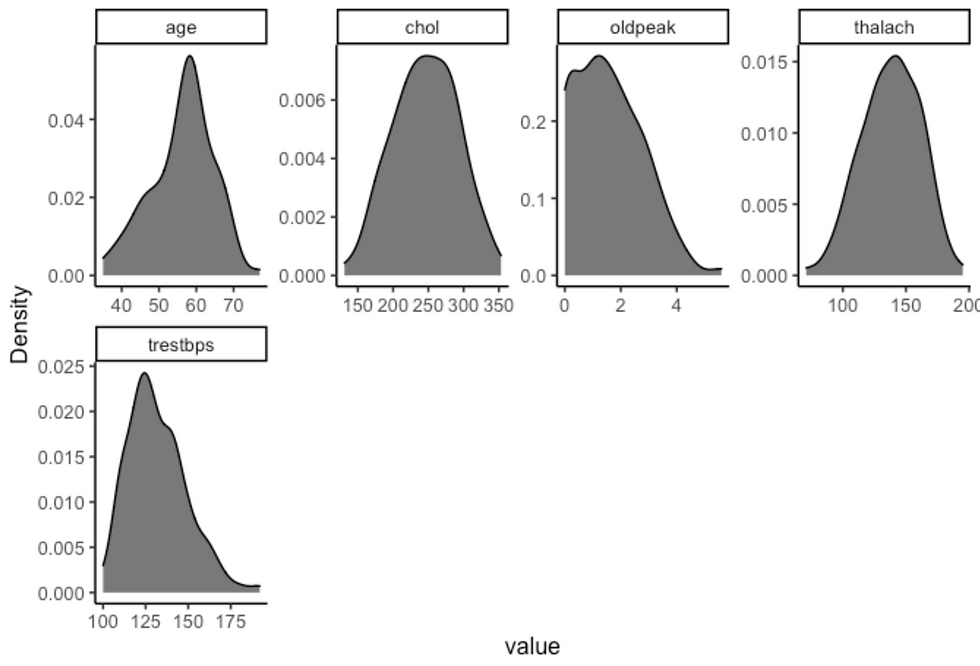
df %>%
filter(sex == "male", target == "asymptomatic") %>%
plot_bar(ggtheme = theme_classic2())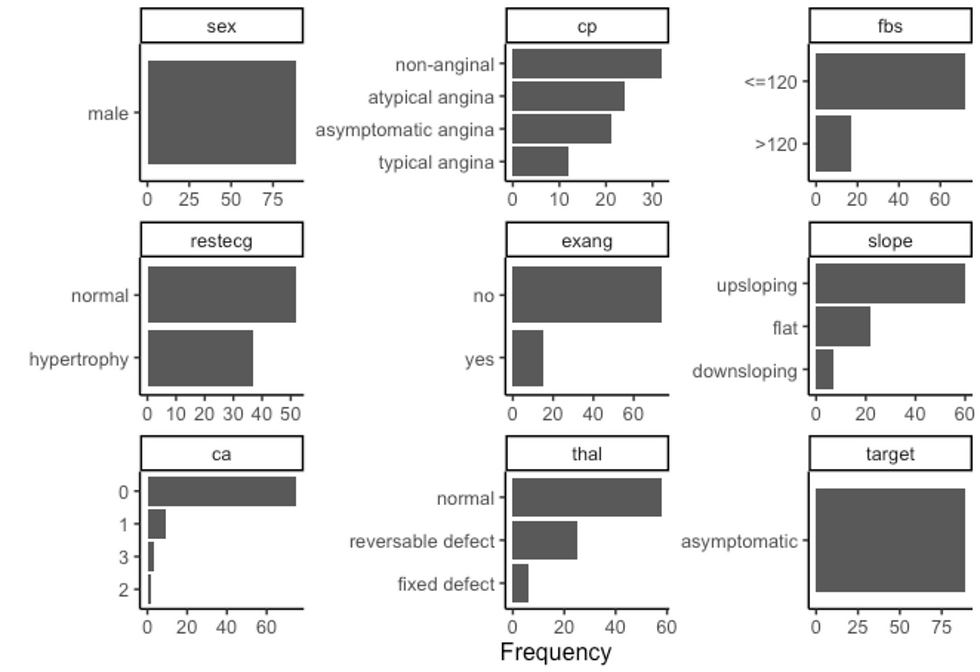
df %>%
filter(sex == "male", target == "heart-disease") %>%
plot_bar(ggtheme = theme_classic2())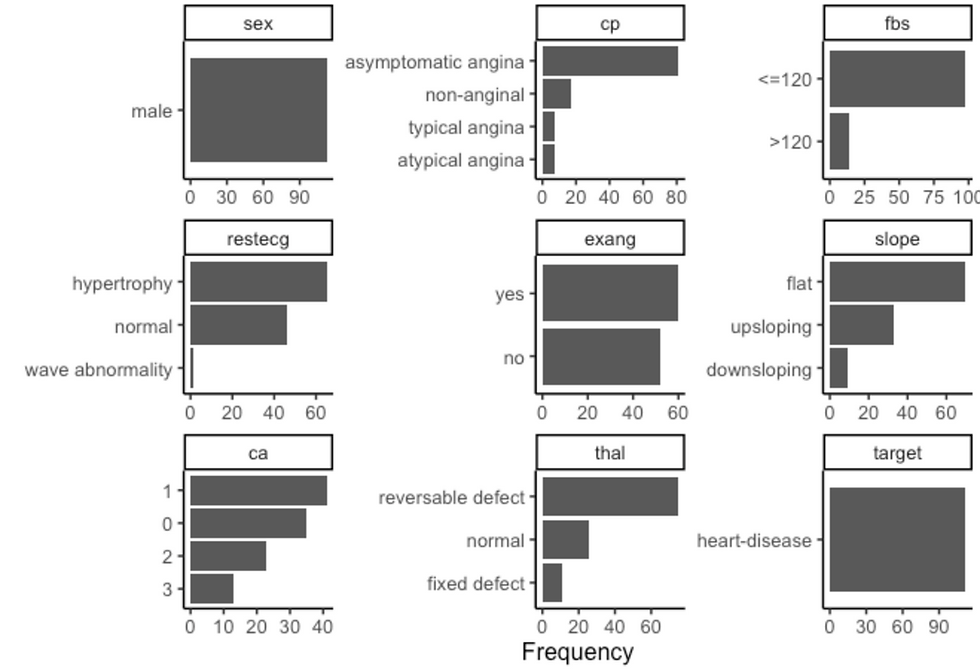
E Prepare a summary table per disease and gender
df %>%
group_by(target, sex) %>%
summarise(
n_disease = n(),
mean_age = round(mean(age), digits=2),
sd_age = round(sd(age), digits=2),
mean_trestbps = round(mean(trestbps), digits=2),
sd_trestbps = round(sd(trestbps), digits=2),
mean_chol = round(mean(chol), digits=2),
sd_chol = round(sd(chol), digits=2),
mean_thalach = round(mean(thalach), digits=2),
sd_thalach = round(sd(thalach), digits=2),
mean_oldpeak = round(mean(oldpeak), digits=2),
sd_oldpeak = round(sd(oldpeak), digits=2)
)## `summarise()` has grouped output by 'target'. You can override using the `.groups` argument.
## # A tibble: 4 x 13
## # Groups: target [2]
## target sex n_disease mean_age sd_age mean_trestbps sd_trestbps mean_chol
## <chr> <fct> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 asymp… fema… 71 54.6 10.3 129. 16.6 257.
## 2 asymp… male 89 51.1 8.63 130. 16.2 232.
## 3 heart… fema… 24 59.0 4.96 146. 21.4 275.
## 4 heart… male 112 56.2 8.36 132. 17.4 246.
## # … with 5 more variables: sd_chol <dbl>, mean_thalach <dbl>, sd_thalach <dbl>,
## # mean_oldpeak <dbl>, sd_oldpeak <dbl>IV Data Visualisation
From the Exploratory Data analysis, it seems that several differences are statistically significant according to gender and health status.
A Visualisation of variables per gender
# Male and Female count
a1 <- ggplot(df, aes(x = sex, fill = sex)) +
geom_bar(width = 0.5) +
scale_fill_manual(values = c("#386cb0","#fdb462"))+
theme_classic2() +
theme(legend.position='none')
# Age per gender
b1 <- ggplot(df, aes(x= sex, y = age, fill = sex)) +
geom_violin(width = 0.5) +
geom_boxplot(width = 0.2) +
ylim(0, 90) +
stat_compare_means(aes(label = ..p.signif..), method = "t.test") +
scale_fill_manual(values = c("#386cb0","#fdb462"))+
theme_classic2() +
theme(legend.position='none')
# trestbps
c1 <- ggplot(df, aes(x = sex, y = trestbps, fill = sex)) +
geom_violin(width = 0.5) +
geom_boxplot(width = 0.2) +
labs(y = "blood pressure (mmHg)") +
ylim(0,250) +
stat_compare_means(aes(label = ..p.signif..), method = "t.test") +
scale_fill_manual(values = c("#386cb0","#fdb462"))+
theme_classic2() +
theme(legend.position='none')
# chol
d1 <- ggplot(df, aes(x = sex, y = chol, fill = sex)) +
geom_violin(width = 0.5) +
geom_boxplot(width = 0.2) +
labs(y = "cholestorol (mg/dl)") +
ylim(0,500) +
stat_compare_means(aes(label = ..p.signif..), method = "t.test") +
scale_fill_manual(values = c("#386cb0","#fdb462"))+
theme_classic2() +
theme(legend.position='none')
# oldpeak
e1 <- ggplot(df, aes(x = sex, y = oldpeak, fill = sex)) +
geom_violin(width = 0.5) +
geom_boxplot(width = 0.2) +
labs(y = "ST depression") +
ylim(0,10) +
stat_compare_means(aes(label = ..p.signif..), method = "t.test") +
scale_fill_manual(values = c("#386cb0","#fdb462"))+
theme_classic2() +
theme(legend.position='none')
# thalach
f1 <- ggplot(df, aes(x = sex, y = thalach, fill = sex)) +
geom_violin(width = 0.5) +
geom_boxplot(width = 0.2) +
labs(y = "Max. heart rate") +
ylim(0,250) +
stat_compare_means(aes(label = ..p.signif..), method = "t.test") +
scale_fill_manual(values = c("#386cb0","#fdb462"))+
theme_classic2() +
theme(legend.position='none')
suppressWarnings(ggarrange(a1, b1, c1, d1, e1, f1,
ncol = 2, nrow = 3,
align = "v"))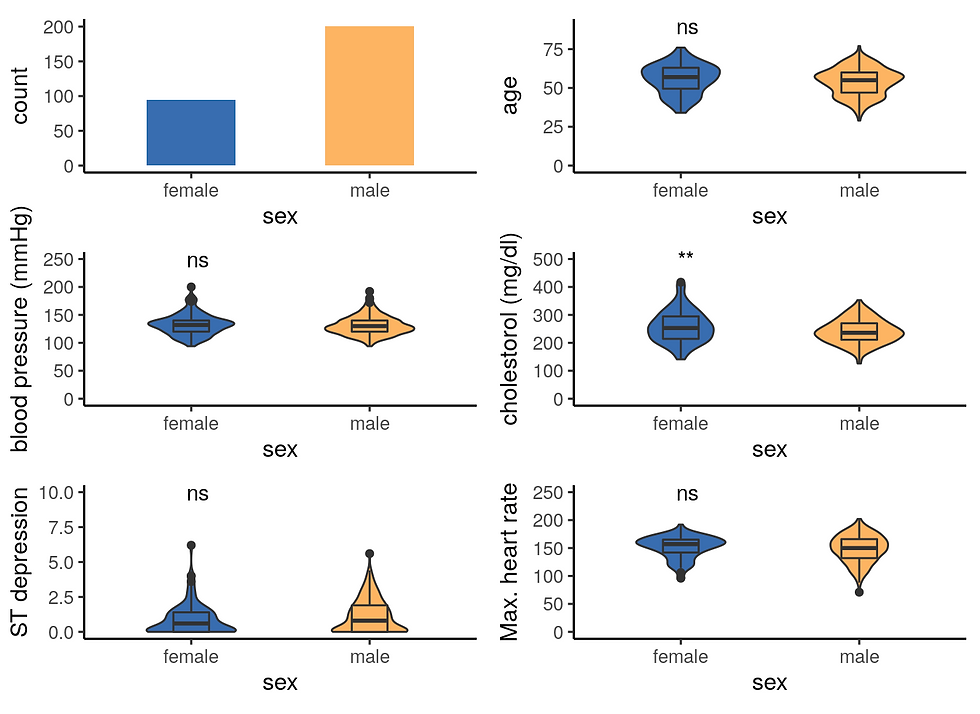
# Disease status
g1 <- ggplot(df, aes(x = target, fill = sex)) +
geom_bar(width = 0.5, position = 'dodge') +
labs(x = "") +
coord_flip() +
scale_fill_manual(values = c("#386cb0","#fdb462"))+
theme_classic2() +
theme(legend.position='none')
# cp
h1 <- ggplot(df, aes(cp, group = sex, fill = sex)) +
geom_bar(position = "dodge") +
labs(x = "", y = "chest pain") +
coord_flip() +
scale_fill_manual(values = c("#386cb0","#fdb462"))+
theme_classic2() +
theme(legend.position='none')
# restecg
i1 <- ggplot(df, aes(restecg, group = sex, fill = sex)) +
geom_bar(position = "dodge") +
labs(x = "", y = "rest. electrocardiographic") +
coord_flip() +
scale_fill_manual(values = c("#386cb0","#fdb462"))+
theme_classic2() +
theme(legend.position='none')
# slope
j1 <- ggplot(df, aes(slope, group = sex, fill = sex)) +
geom_bar(position = "dodge") +
labs(x = "", y = "peak exercise ST") +
coord_flip() +
scale_fill_manual(values = c("#386cb0","#fdb462"))+
theme_classic2() +
theme(legend.position='none')
# thal
k1 <- ggplot(df, aes(thal, group = sex, fill = sex)) +
geom_bar(position = "dodge") +
labs(x = "", y = "Thalium stress test") +
coord_flip() +
scale_fill_manual(values = c("#386cb0","#fdb462"))+
theme_classic2() +
theme(legend.position='none')
# fbp
l1 <- ggplot(df, aes(fbs, group = sex, fill = sex)) +
geom_bar(position = "dodge") +
labs(x = "", y = "Fasting blood sugar") +
coord_flip() +
scale_fill_manual(values = c("#386cb0","#fdb462"))+
theme_classic2() +
theme(legend.position='none')
# exang
m1 <- ggplot(df, aes(exang, group = sex, fill = sex)) +
geom_bar(position = "dodge") +
labs(x = "", y = "Exercise induced angina") +
coord_flip() +
scale_fill_manual(values = c("#386cb0","#fdb462"))+
theme_classic2() +
theme(legend.position='none')
# ca
n1 <- ggplot(df, aes(ca, group = sex, fill = sex)) +
geom_bar(position = "dodge") +
labs(x = "", y = "flourosopy") +
coord_flip() +
scale_fill_manual(values = c("#386cb0","#fdb462"))+
theme_classic2() +
theme(legend.position='none')
ggarrange(g1, h1, i1, j1, k1, l1, m1, n1,
ncol = 2, nrow = 4,
align = "v")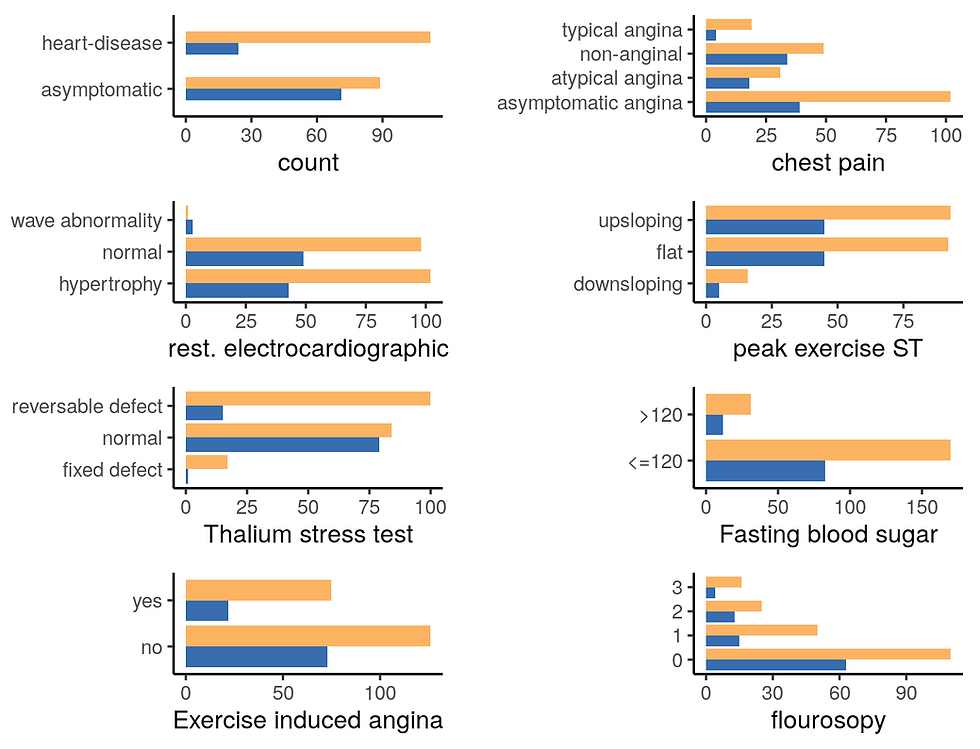
From this first plot, it appears that this dataset contains more males patients with a higher proportion of heart disease compared to female patients.
B Visualisation of variables per disease status
1 Male patient
df <- df2 %>%
filter(sex == "male")
# Male and Female count
a2 <- ggplot(df, aes(x = target, fill = target)) +
geom_bar(width = 0.5, position = 'dodge') +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# Age per gender
b2 <- ggplot(df, aes(x= target, y = age, fill = target)) +
geom_violin(width = 0.5) +
geom_boxplot(width = 0.2) +
ylim(0, 90) +
stat_compare_means(aes(label = ..p.signif..), method = "t.test") +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# trestbps
c2 <- ggplot(df, aes(x = target, y = trestbps, fill = target)) +
geom_violin(width = 0.5) +
geom_boxplot(width = 0.2) +
labs(y = "blood pressure (mmHg)") +
ylim(0,250) +
stat_compare_means(aes(label = ..p.signif..), method = "t.test") +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# chol
d2 <- ggplot(df, aes(x = target, y = chol, fill = target)) +
geom_violin(width = 0.5) +
geom_boxplot(width = 0.2) +
labs(y = "cholestorol (mg/dl)") +
ylim(0,500) +
stat_compare_means(aes(label = ..p.signif..), method = "t.test") +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# oldpeak
e2 <- ggplot(df, aes(x = target, y = oldpeak, fill = target)) +
geom_violin(width = 0.5) +
geom_boxplot(width = 0.2) +
labs(y = "ST depression") +
ylim(0,10) +
stat_compare_means(aes(label = ..p.signif..), method = "t.test") +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# thalach
f2 <- ggplot(df, aes(x = target, y = thalach, fill = target)) +
geom_violin(width = 0.5) +
geom_boxplot(width = 0.2) +
labs(y = "Max. heart rate") +
ylim(0,250) +
stat_compare_means(aes(label = ..p.signif..), method = "t.test") +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
ggarrange(a2, b2, c2, d2, e2, f2,
ncol = 2, nrow = 3,
align = "v")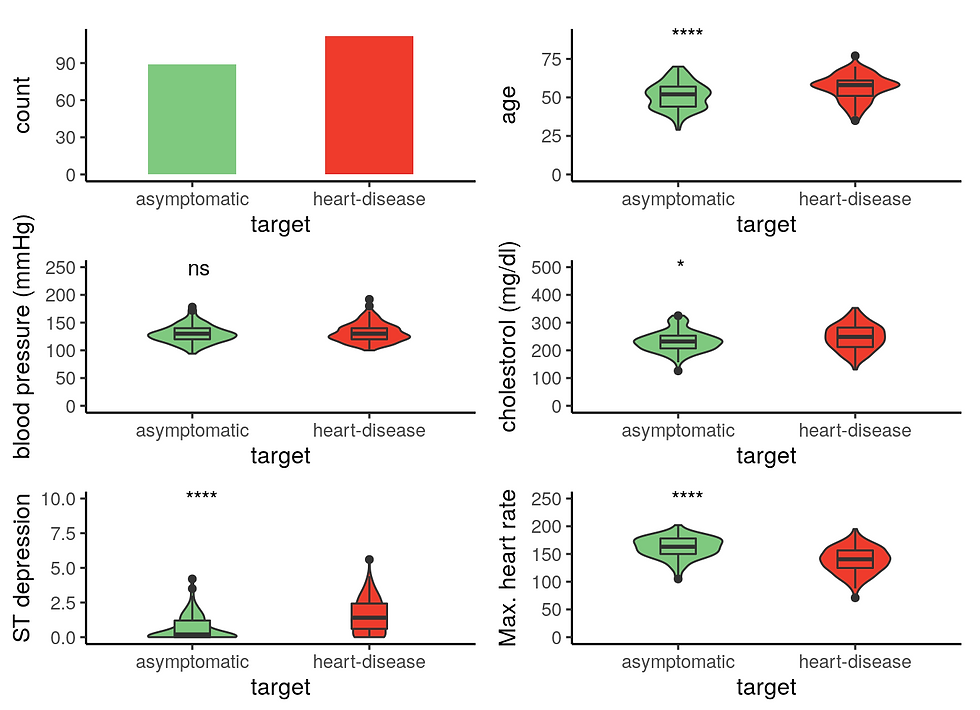
Male patients with heart disease are significantly older, have higher cholesterol level, and reduced maximum heart rate response to the thallium test.
# Disease status
g2 <- ggplot(df, aes(x = target, fill = target)) +
geom_bar(width = 0.5, position = 'dodge') +
labs(x = "") +
coord_flip() +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# cp
h2 <- ggplot(df, aes(cp, group = target, fill = target)) +
geom_bar(position = "dodge") +
labs(x = "", y = "chest pain") +
coord_flip() +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# restecg
i2 <- ggplot(df, aes(restecg, group = target, fill = target)) +
geom_bar(position = "dodge") +
labs(x = "", y = "rest. electrocardiographic") +
coord_flip() +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# slope
j2 <- ggplot(df, aes(slope, group = target, fill = target)) +
geom_bar(position = "dodge") +
labs(x = "", y = "peak exercise ST") +
coord_flip() +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# thal
k2 <- ggplot(df, aes(thal, group = target, fill = target)) +
geom_bar(position = "dodge") +
labs(x = "", y = "Thalium stress test") +
coord_flip() +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# fbp
l2 <- ggplot(df, aes(fbs, group = target, fill = target)) +
geom_bar(position = "dodge") +
labs(x = "", y = "Fasting blood sugar") +
coord_flip() +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# exang
m2 <- ggplot(df, aes(exang, group = target, fill = target)) +
geom_bar(position = "dodge") +
labs(x = "", y = "Exercise induced angina") +
coord_flip() +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# ca
n2 <- ggplot(df, aes(ca, group = target, fill = target)) +
geom_bar(position = "dodge") +
labs(x = "", y = "flourosopy") +
coord_flip() +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
ggarrange(g2, h2, i2, j2, k2, l2, m2, n2,
ncol = 2, nrow = 4,
align = "v")
2 Female patients
df <- df2 %>%
filter(sex == "female")
# Male and Female count
a2 <- ggplot(df, aes(x = target, fill = target)) +
geom_bar(width = 0.5, position = 'dodge') +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# Age per gender
b2 <- ggplot(df, aes(x= target, y = age, fill = target)) +
geom_violin(width = 0.5) +
geom_boxplot(width = 0.2) +
ylim(0, 90) +
stat_compare_means(aes(label = ..p.signif..), method = "t.test") +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# trestbps
c2 <- ggplot(df, aes(x = target, y = trestbps, fill = target)) +
geom_violin(width = 0.5) +
geom_boxplot(width = 0.2) +
labs(y = "blood pressure (mmHg)") +
ylim(0,250) +
stat_compare_means(aes(label = ..p.signif..), method = "t.test") +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# chol
d2 <- ggplot(df, aes(x = target, y = chol, fill = target)) +
geom_violin(width = 0.5) +
geom_boxplot(width = 0.2) +
labs(y = "cholestorol (mg/dl)") +
ylim(0,500) +
stat_compare_means(aes(label = ..p.signif..), method = "t.test") +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# oldpeak
e2 <- ggplot(df, aes(x = target, y = oldpeak, fill = target)) +
geom_violin(width = 0.5) +
geom_boxplot(width = 0.2) +
labs(y = "ST depression") +
ylim(0,10) +
stat_compare_means(aes(label = ..p.signif..), method = "t.test") +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# thalach
f2 <- ggplot(df, aes(x = target, y = thalach, fill = target)) +
geom_violin(width = 0.5) +
geom_boxplot(width = 0.2) +
labs(y = "Max. heart rate") +
ylim(0,250) +
stat_compare_means(aes(label = ..p.signif..), method = "t.test") +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
suppressWarnings(ggarrange(a2, b2, c2, d2, e2, f2,
ncol = 2, nrow = 3,
align = "v"))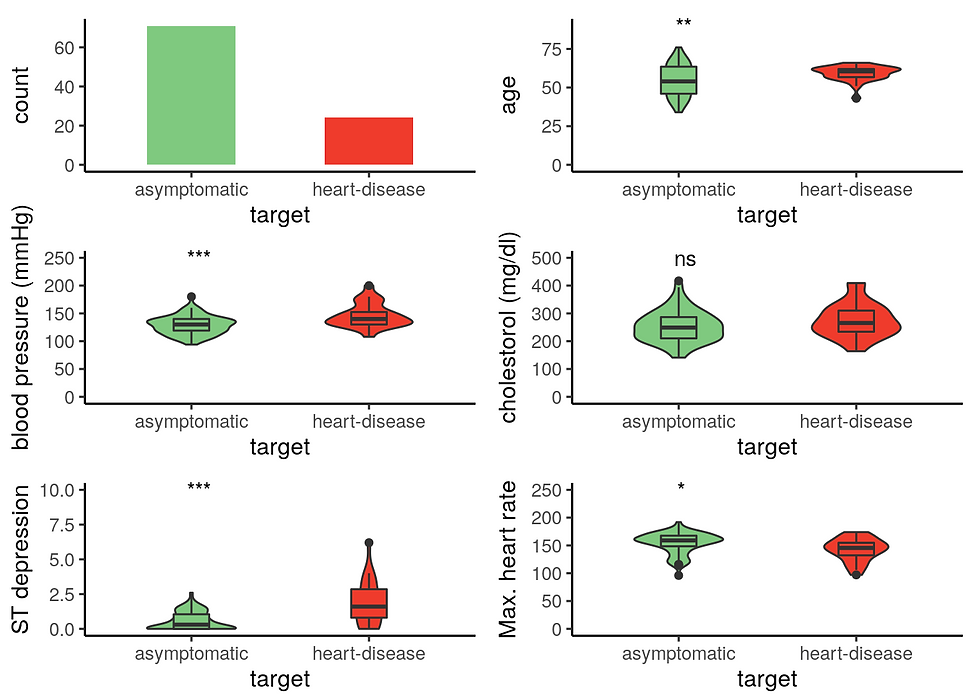
There is less woman with heart disease on this data set. Women with heart disease have a significantly higher resting blood presure contrary to male with heart disease. Similarly to men, women with heart disease have a lower maximum heart rate in response to the thallium test.
# Disease status
g2 <- ggplot(df, aes(x = target, fill = target)) +
geom_bar(width = 0.5, position = 'dodge') +
labs(x = "") +
coord_flip() +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# cp
h2 <- ggplot(df, aes(cp, group = target, fill = target)) +
geom_bar(position = "dodge") +
labs(x = "", y = "chest pain") +
coord_flip() +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# restecg
i2 <- ggplot(df, aes(restecg, group = target, fill = target)) +
geom_bar(position = "dodge") +
labs(x = "", y = "rest. electrocardiographic") +
coord_flip() +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# slope
j2 <- ggplot(df, aes(slope, group = target, fill = target)) +
geom_bar(position = "dodge") +
labs(x = "", y = "peak exercise ST") +
coord_flip() +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# thal
k2 <- ggplot(df, aes(thal, group = target, fill = target)) +
geom_bar(position = "dodge") +
labs(x = "", y = "Thalium stress test") +
coord_flip() +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# fbp
l2 <- ggplot(df, aes(fbs, group = target, fill = target)) +
geom_bar(position = "dodge") +
labs(x = "", y = "Fasting blood sugar") +
coord_flip() +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# exang
m2 <- ggplot(df, aes(exang, group = target, fill = target)) +
geom_bar(position = "dodge") +
labs(x = "", y = "Exercise induced angina") +
coord_flip() +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
# ca
n2 <- ggplot(df, aes(ca, group = target, fill = target)) +
geom_bar(position = "dodge") +
labs(x = "", y = "flourosopy") +
coord_flip() +
scale_fill_manual(values = c("#7fc97f","#ef3b2c"))+
theme_classic2() +
theme(legend.position='none')
ggarrange(g2, h2, i2, j2, k2, l2, m2, n2,
ncol = 2, nrow = 4,
align = "v")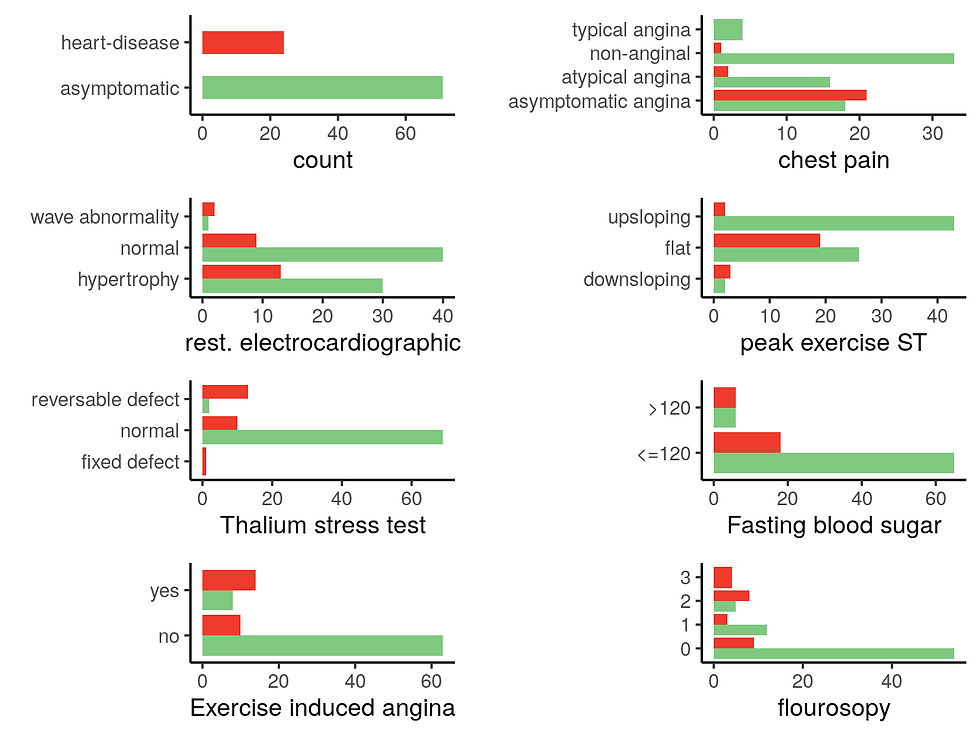
V Correlations
A use the numerical dataset
df <- copy %>%
filter(
thal != 0 & ca != 4 # remove values corresponding to NA in original dataset
)
# ggcorr(df, palette = "RdBu")
GGally::ggcorr(df, geom = "circle")
select2 <- df %>%
dplyr::select(
target,
slope,
thalach,
restecg,
cp
)
ggcorr(select2, geom = "circle")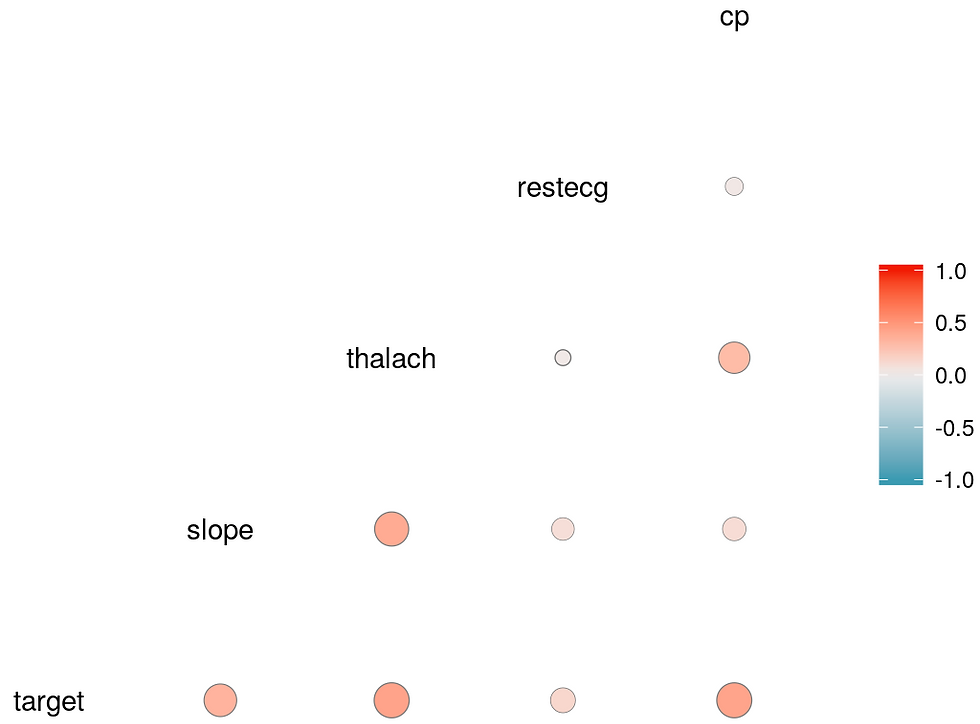
ggpairs(select2)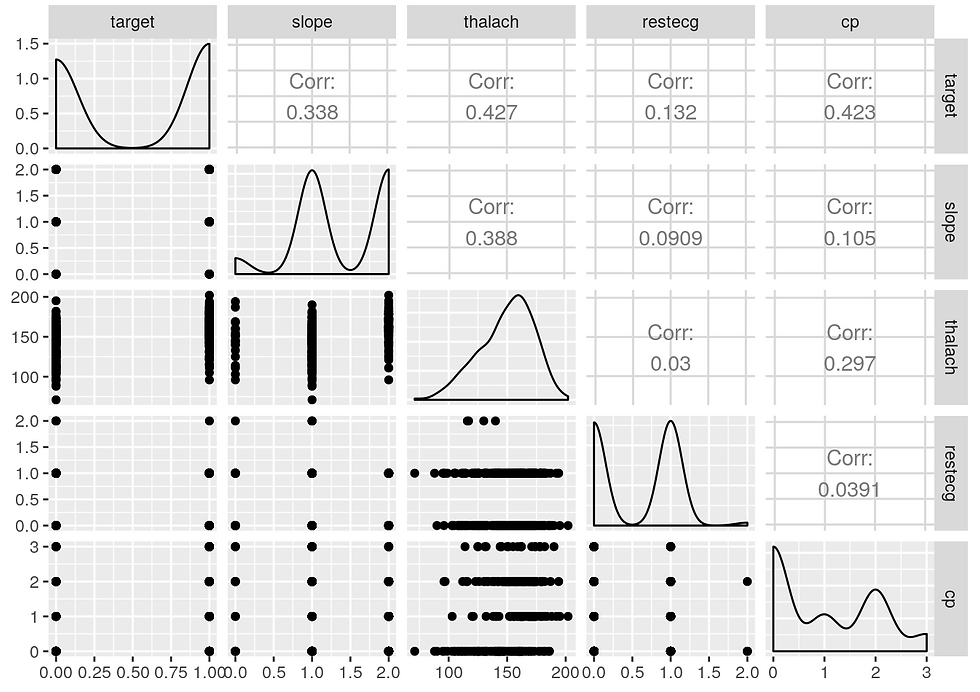
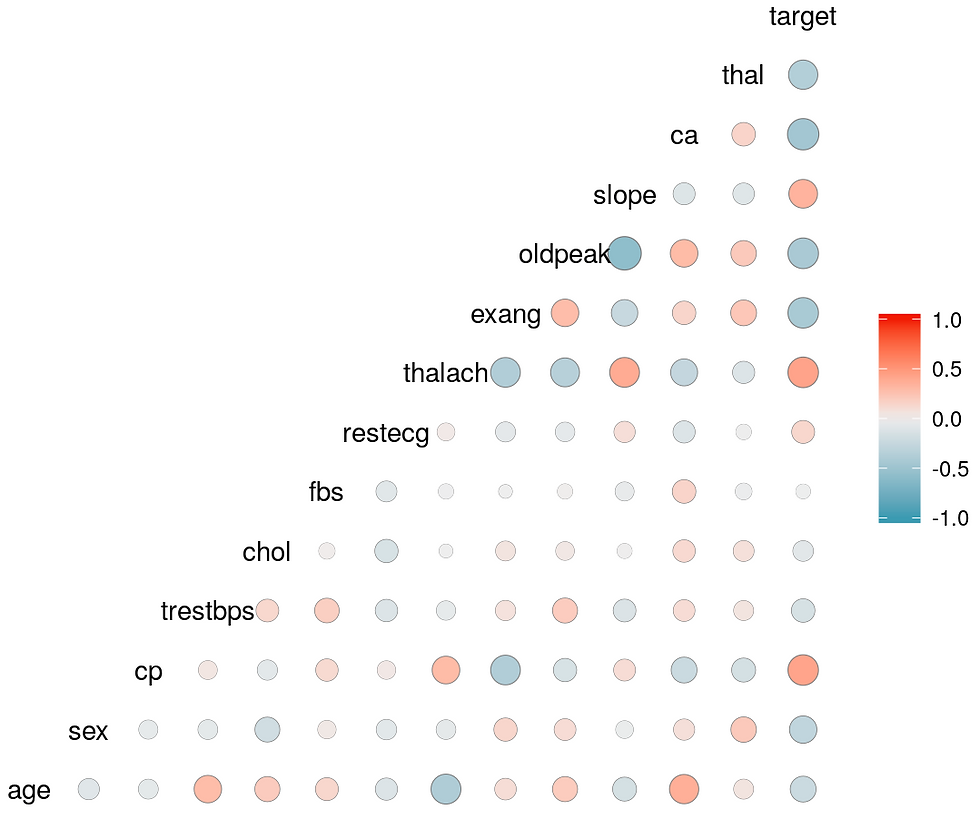
From the correlation study it seems that the parameters: cp, restecg, thalach, slope
are the most useful to predict the risk of heart disease
From the EDA analysis it seems that: age, sex, cholesterol, restecg
are also useful
For prediction the following variables seems the most useful: age, sex, cholesterol, restecg, cp, thalach, slope
VI Machine Learning: classification model with rpart and random forest packages
Select the columns useful for prediction according to the EDA analysis.
Separate the data set in a train and test subsets.
Build a classification tree model with rpart.
Print model accuracy and decision tree.
A Use select columns for classification
df_select <- df %>%
dplyr::select( #because of conflict between MASS and dplyr select need to use dplyr::select
target,
age,
sex,
chol,
restecg,
cp,
thalach,
slope
)
df_select$target <- factor(df_select$target) # Define target as a factor. rpart classification would not work otherwise.
accuracy <- 0
# Build a simple classification desicion tree with rpart. Run the model until the accuracy reach the selected minimum.
while(accuracy <= 0.85) {
split_values <- sample.split(df_select$target, SplitRatio = 0.65)
train_set <- subset(df_select, split_values == T)
test_set <- subset(df_select, split_values == F)
mod_class <- rpart(target~. , data=train_set)
result_class <- predict(mod_class, test_set, type = "class")
table <- table(test_set$target, result_class)
accuracy <- (table["0","0"] + table["1","1"])/sum(table)
# cat("accuracy = ", round(accuracy, digits = 2)*100, "%")
}Print model accuracy.
According to this parameters the model should be at least 85% accurate.
cat("Model accuracy", round(accuracy, digits = 2)*100, "%") ## Model accuracy 86 %Print the decision tree.
# par(mfrow = c(1,2), xpd = NA) # otherwise on some devices the text is clipped
fancyRpartPlot(mod_class, , caption = NULL)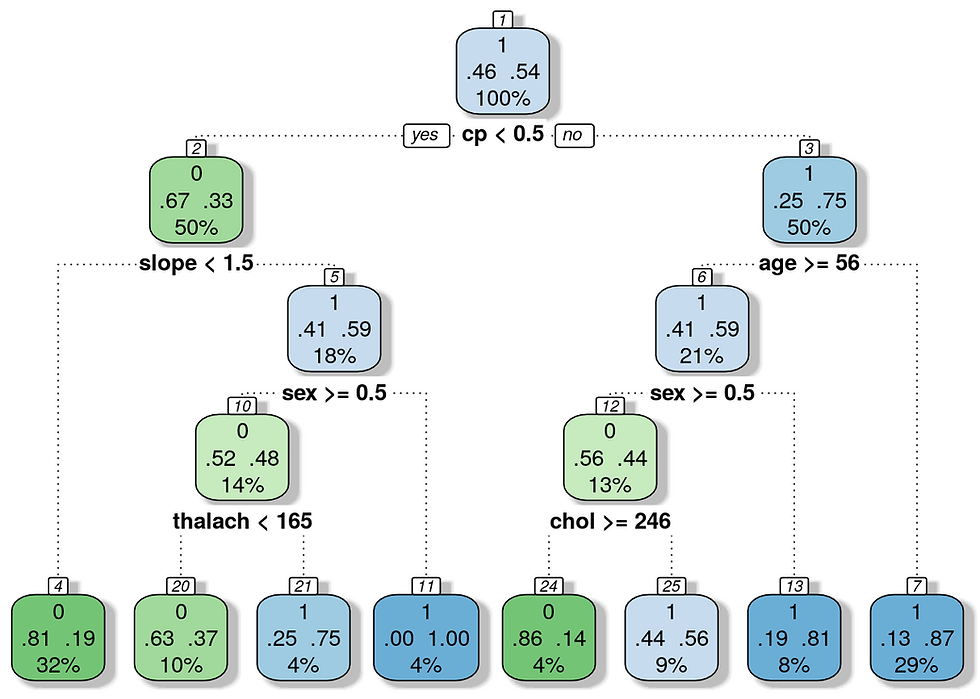
B Use the full dataset for classification
copy2 <- df
df$target <- factor(df$target)
accuracy <- 0
# Build a simple classification desicion tree with rpart. Run the model until the accuracy reach the selected minimum.
while(accuracy <= 0.88) {
split_values <- sample.split(df_select$target, SplitRatio = 0.65)
train_set <- subset(df, split_values == T)
test_set <- subset(df, split_values == F)
mod_class <- rpart(target~. , data=train_set)
result_class <- predict(mod_class, test_set, type = "class")
table <- table(test_set$target, result_class)
accuracy <- (table["0","0"] + table["1","1"])/sum(table)
# cat("accuracy = ", round(accuracy, digits = 2)*100, "%")
}Print model accuracy.
According to the parameters the model should be at least 88% accurate.
cat("Model accuracy", round(accuracy, digits = 2)*100, "%") ## Model accuracy 89 %Print the decision tree.
# par(mfrow = c(1,2), xpd = NA) # otherwise on some devices the text is clipped
fancyRpartPlot(mod_class, , caption = NULL)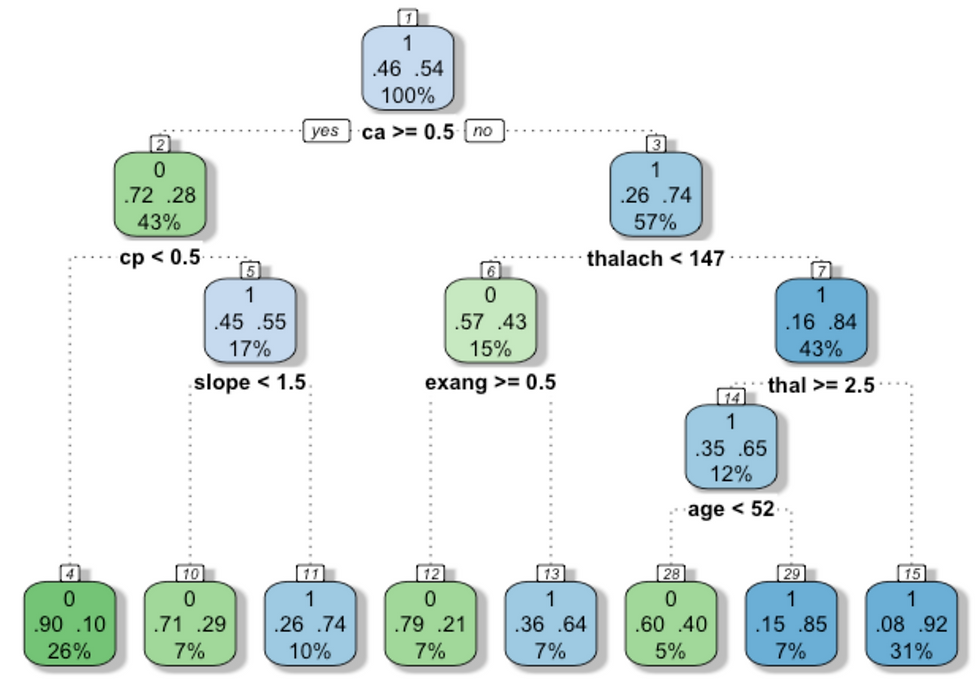
C Prediction on selected column with random forest
set.seed(123)
train <- sample(nrow(df_select), 0.7*nrow(df_select), replace = FALSE)
TrainSet <- df_select[train,]
ValidSet <- df_select[-train,]
summary(TrainSet)## target age sex chol restecg
## 0: 91 Min. :29.00 Min. :0.0000 Min. :126 Min. :0.0000
## 1:116 1st Qu.:48.00 1st Qu.:0.0000 1st Qu.:208 1st Qu.:0.0000
## Median :56.00 Median :1.0000 Median :240 Median :0.0000
## Mean :55.02 Mean :0.6763 Mean :244 Mean :0.5024
## 3rd Qu.:62.00 3rd Qu.:1.0000 3rd Qu.:272 3rd Qu.:1.0000
## Max. :77.00 Max. :1.0000 Max. :564 Max. :2.0000
## cp thalach slope
## Min. :0.000 Min. : 95.0 Min. :0.000
## 1st Qu.:0.000 1st Qu.:136.0 1st Qu.:1.000
## Median :1.000 Median :153.0 Median :1.000
## Mean :1.014 Mean :150.2 Mean :1.386
## 3rd Qu.:2.000 3rd Qu.:165.0 3rd Qu.:2.000
## Max. :3.000 Max. :202.0 Max. :2.000summary(ValidSet)## target age sex chol restecg
## 0:45 Min. :35.00 Min. :0.0000 Min. :175.0 Min. :0.000
## 1:44 1st Qu.:47.00 1st Qu.:0.0000 1st Qu.:223.0 1st Qu.:0.000
## Median :54.00 Median :1.0000 Median :247.0 Median :1.000
## Mean :53.37 Mean :0.6854 Mean :254.5 Mean :0.573
## 3rd Qu.:59.00 3rd Qu.:1.0000 3rd Qu.:283.0 3rd Qu.:1.000
## Max. :70.00 Max. :1.0000 Max. :409.0 Max. :2.000
## cp thalach slope
## Min. :0.0000 Min. : 71.0 Min. :0.000
## 1st Qu.:0.0000 1st Qu.:132.0 1st Qu.:1.000
## Median :0.0000 Median :152.0 Median :1.000
## Mean :0.8315 Mean :148.2 Mean :1.416
## 3rd Qu.:2.0000 3rd Qu.:168.0 3rd Qu.:2.000
## Max. :3.0000 Max. :190.0 Max. :2.000# Create a Random Forest model with default parameters
model1 <- randomForest(target ~ ., data = TrainSet, ntree = 1000, mtry = 1, importance = TRUE)
model1##
## Call:
## randomForest(formula = target ~ ., data = TrainSet, ntree = 1000, mtry = 1, importance = TRUE)
## Type of random forest: classification
## Number of trees: 1000
## No. of variables tried at each split: 1
##
## OOB estimate of error rate: 28.02%
## Confusion matrix:
## 0 1 class.error
## 0 58 33 0.3626374
## 1 25 91 0.2155172# Predicting on Validation set
predValid <- predict(model1, ValidSet, type = "class")
# Checking classification accuracy
mean(predValid == ValidSet$target) ## [1] 0.7977528table(predValid,ValidSet$target)##
## predValid 0 1
## 0 36 9
## 1 9 35# To check important variables
importance(model1) ## 0 1 MeanDecreaseAccuracy MeanDecreaseGini
## age 3.456978 11.118814352 10.045334 10.174882
## sex 17.986950 18.447097660 22.356657 6.212641
## chol -3.667658 -0.005187581 -2.378170 9.368247
## restecg 1.926634 3.951377314 4.138125 3.080821
## cp 19.676722 17.420672992 22.537720 10.408636
## thalach 14.987882 12.899451821 18.350082 14.006896
## slope 13.982290 9.048798817 15.232708 6.022085varImpPlot(model1) 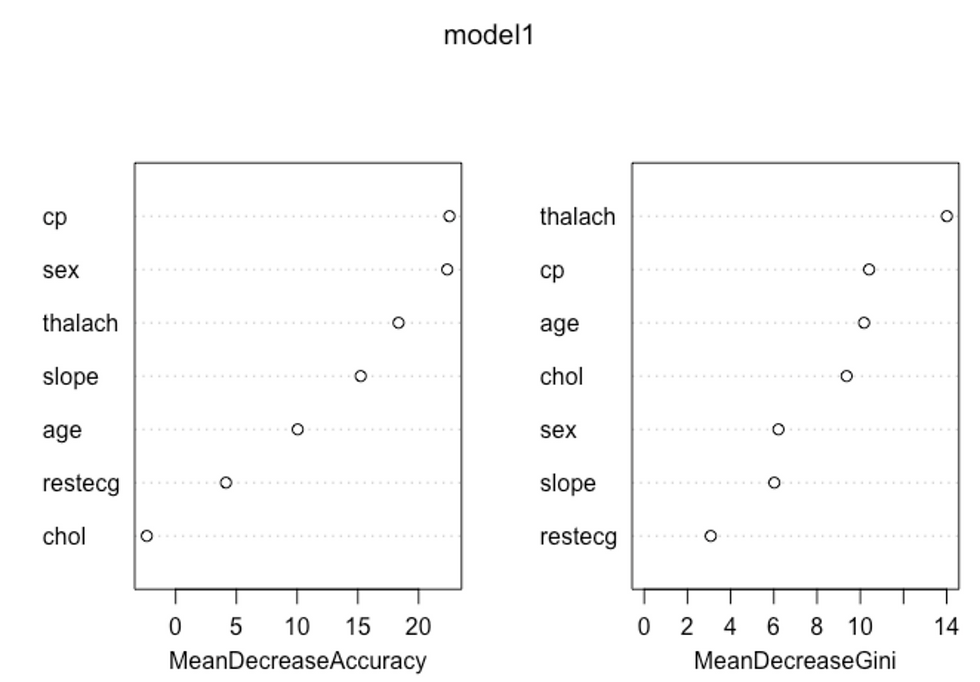
D Use the full dataset for classification with random forest
set.seed(123)
train <- sample(nrow(df), 0.7*nrow(df_select), replace = FALSE)
TrainSet <- df[train,]
ValidSet <- df[-train,]
summary(TrainSet)## age sex cp trestbps chol
## Min. :29.00 Min. :0.0000 Min. :0.000 Min. :100.0 Min. :126
## 1st Qu.:48.00 1st Qu.:0.0000 1st Qu.:0.000 1st Qu.:120.0 1st Qu.:208
## Median :56.00 Median :1.0000 Median :1.000 Median :130.0 Median :240
## Mean :55.02 Mean :0.6763 Mean :1.014 Mean :131.1 Mean :244
## 3rd Qu.:62.00 3rd Qu.:1.0000 3rd Qu.:2.000 3rd Qu.:140.0 3rd Qu.:272
## Max. :77.00 Max. :1.0000 Max. :3.000 Max. :192.0 Max. :564
## fbs restecg thalach exang
## Min. :0.0000 Min. :0.0000 Min. : 95.0 Min. :0.0000
## 1st Qu.:0.0000 1st Qu.:0.0000 1st Qu.:136.0 1st Qu.:0.0000
## Median :0.0000 Median :0.0000 Median :153.0 Median :0.0000
## Mean :0.1643 Mean :0.5024 Mean :150.2 Mean :0.3382
## 3rd Qu.:0.0000 3rd Qu.:1.0000 3rd Qu.:165.0 3rd Qu.:1.0000
## Max. :1.0000 Max. :2.0000 Max. :202.0 Max. :1.0000
## oldpeak slope ca thal target
## Min. :0.000 Min. :0.000 Min. :0.0000 Min. :1.000 0: 91
## 1st Qu.:0.000 1st Qu.:1.000 1st Qu.:0.0000 1st Qu.:2.000 1:116
## Median :0.800 Median :1.000 Median :0.0000 Median :2.000
## Mean :1.055 Mean :1.386 Mean :0.6039 Mean :2.319
## 3rd Qu.:1.600 3rd Qu.:2.000 3rd Qu.:1.0000 3rd Qu.:3.000
## Max. :6.200 Max. :2.000 Max. :3.0000 Max. :3.000summary(ValidSet)## age sex cp trestbps
## Min. :35.00 Min. :0.0000 Min. :0.0000 Min. : 94.0
## 1st Qu.:47.00 1st Qu.:0.0000 1st Qu.:0.0000 1st Qu.:120.0
## Median :54.00 Median :1.0000 Median :0.0000 Median :130.0
## Mean :53.37 Mean :0.6854 Mean :0.8315 Mean :132.8
## 3rd Qu.:59.00 3rd Qu.:1.0000 3rd Qu.:2.0000 3rd Qu.:146.0
## Max. :70.00 Max. :1.0000 Max. :3.0000 Max. :200.0
## chol fbs restecg thalach
## Min. :175.0 Min. :0.0000 Min. :0.000 Min. : 71.0
## 1st Qu.:223.0 1st Qu.:0.0000 1st Qu.:0.000 1st Qu.:132.0
## Median :247.0 Median :0.0000 Median :1.000 Median :152.0
## Mean :254.5 Mean :0.1011 Mean :0.573 Mean :148.2
## 3rd Qu.:283.0 3rd Qu.:0.0000 3rd Qu.:1.000 3rd Qu.:168.0
## Max. :409.0 Max. :1.0000 Max. :2.000 Max. :190.0
## exang oldpeak slope ca
## Min. :0.0000 Min. :0.000 Min. :0.000 Min. :0.0000
## 1st Qu.:0.0000 1st Qu.:0.000 1st Qu.:1.000 1st Qu.:0.0000
## Median :0.0000 Median :0.800 Median :1.000 Median :1.0000
## Mean :0.3034 Mean :1.069 Mean :1.416 Mean :0.8539
## 3rd Qu.:1.0000 3rd Qu.:1.900 3rd Qu.:2.000 3rd Qu.:1.0000
## Max. :1.0000 Max. :4.400 Max. :2.000 Max. :3.0000
## thal target
## Min. :1.000 0:45
## 1st Qu.:2.000 1:44
## Median :2.000
## Mean :2.348
## 3rd Qu.:3.000
## Max. :3.000# Create a Random Forest model with default parameters
model2 <- randomForest(target ~ ., data = TrainSet, ntree = 1000, mtry = 2, importance = TRUE)
model2##
## Call:
## randomForest(formula = target ~ ., data = TrainSet, ntree = 1000, mtry = 2, importance = TRUE)
## Type of random forest: classification
## Number of trees: 1000
## No. of variables tried at each split: 2
##
## OOB estimate of error rate: 20.77%
## Confusion matrix:
## 0 1 class.error
## 0 63 28 0.3076923
## 1 15 101 0.1293103# Predicting on train set
predTrain <- predict(model2, TrainSet, type = "class")
# Checking classification accuracy
table(predTrain, TrainSet$target)##
## predTrain 0 1
## 0 91 0
## 1 0 116# Predicting on Validation set
predValid <- predict(model2, ValidSet, type = "class")
# Checking classification accuracy
mean(predValid == ValidSet$target)
## [1] 0.8764045table(predValid,ValidSet$target)##
## predValid 0 1
## 0 39 5
## 1 6 39# To check important variables
importance(model2)
## 0 1 MeanDecreaseAccuracy MeanDecreaseGini
## age 4.050036 6.7463054 7.778196 8.939681
## sex 8.638348 14.3957620 15.929672 4.589083
## cp 15.051396 11.2883097 17.483834 9.059950
## trestbps 1.813027 0.9226925 2.032148 7.050590
## chol -1.726460 -2.4182808 -2.982013 8.319708
## fbs -1.802818 4.1916949 2.098540 1.367634
## restecg 1.178421 2.2335381 2.450169 2.609878
## thalach 9.766391 10.4880323 14.128124 12.069991
## exang 9.185204 5.9611054 10.635688 4.988638
## oldpeak 18.583582 18.0805610 24.266457 12.847074
## slope 8.688542 4.0812821 9.073868 4.321699
## ca 22.053674 24.4175996 29.812574 11.204931
## thal 14.724654 16.1138529 19.564541 8.636882varImpPlot(model2)
References
Data transformation
Kaggles notebooks:
Data Processing
for Machine Learning
https://www.kaggle.com/naik170106027/prediction-of-heart-diseases
https://www.kaggle.com/ekrembayar/heart-disease-uci-eda-models-with-r
https://www.kaggle.com/anirbanshaw24/heart-disease-prediction-and-indicators
https://www.youtube.com/watch?v=SeyghJ5cdm4&feature=youtu.be
https://www.rdocumentation.org/packages/rpart/versions/4.1-15/topics/rpart
https://stackoverflow.com/questions/33767804/invalid-prediction-for-rpart-object-error
https://machinelearningmastery.com/overfitting-and-underfitting-with-machine-learning-algorithms/
https://www.gormanalysis.com/blog/decision-trees-in-r-using-rpart/
https://www.kaggle.com/wguesdon/tuning-random-forest-parameters/edit



Comments